Clustering: Affinity Propagation
Affinity Propagation Introduction
Developed recently (2007), a centroid based clustering algorithm similar to k Means or K medoids
Affinity propagation finds "exemplars" i.e. members of the input set that are representative of clusters.
It uses a graph based approach to let points 'vote' on their preferred 'exemplar'. The end result is a set of cluster 'exemplars' from which we derive clusters by essentially doing what K-Means does and assigning each point to the cluster of it's nearest exemplar.
We need to calculate the following matrices:
Here we must specify to notation:= row, = column - Similarity matrix
- Responsibility matrix
- Availability matrix
- Criterion matrix
Similarity matrix
- Rationale: information about the similarity between any instances, for an element i we look for another element j for which
is the highest (least negative). Hence the diagonal values are all set to the most negative to exclude the case where i find i itself - Barring those on the diagonal, every cell in the similarity matrix is calculated by the negative sum of the squares differences between participants.
- Note that the diagonal values will not be just 0: It is
- Rationale: information about the similarity between any instances, for an element i we look for another element j for which
Responsibility matrix
Rationale:
quantifies how well-suited element k is, to be an exemplar for element i , taking into account the nearest contender k’ to be an exemplar for i. We initialize R matrix with zeros.
Then calculate every cell in the responsibility matrix using the following formula:
- Interpretation: R_{i,k} can be thought of as relative similarity between i and k. It quantifies how similar is i to k, compared to some k’, taking into account the availability of k’. The responsibility of k towards i will decrease as the availability of some other k’ to i increases.
Availability matrix
- Rationale: It quantifies how appropriate is it for i to choose k as its exemplar, taking into account the support from other elements that k should an exemplar.
- The Availability formula for different instances is
- The Self-Availability is
- Interpretation of the formulas
- Availability is self-responsibility of k plus the positive responsibilities of k towards elements other than i.
- We include only positive responsibilities as an exemplar should be positively responsible/explain at least for some data points well, regardless of how poorly it explains other data points.
- If self-responsibility is negative, it means that k is more suitable to belong to another exemplar, rather than being an exemplar. The maximum value of
is 0. reflects accumulated evidence that point k is suitable to be an exemplar, based on the positive responsibilities of k towards other elements.
and matrices are iteratively updated. This procedure may be terminated after a fixed number of iterations, after changes in the values obtained fall below a threshold, or after the values stay constant for some number of iterations. Criterion Matrix
- Criterion matrix is calculated after the updating is terminated.
- Criterion matrix
is the sum of and : - An element i will be assigned to an exemplar k which is not only highly responsible but also highly available to i.
- The highest criterion value of each row is designated as the exemplar. Rows that share the same exemplar are in the same cluster.
Sample run
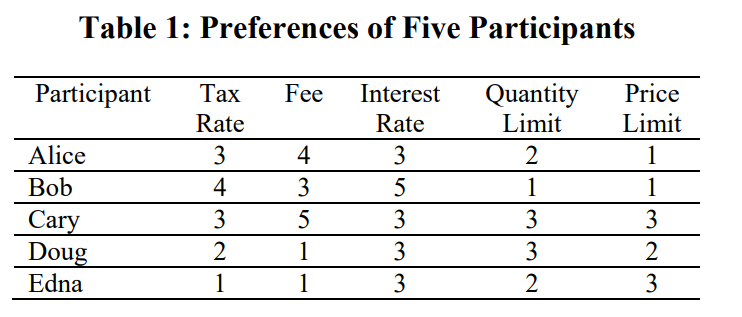
Data 
Similarity Matrix 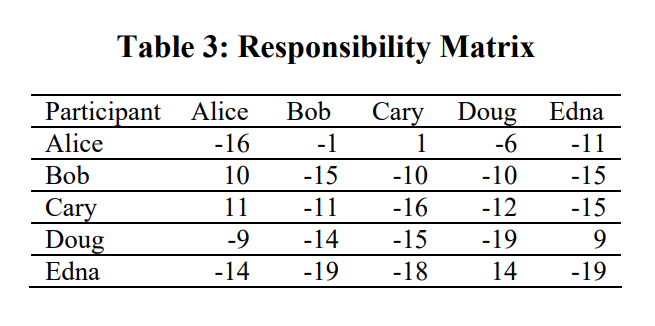
Responsibility Matrix (First round) 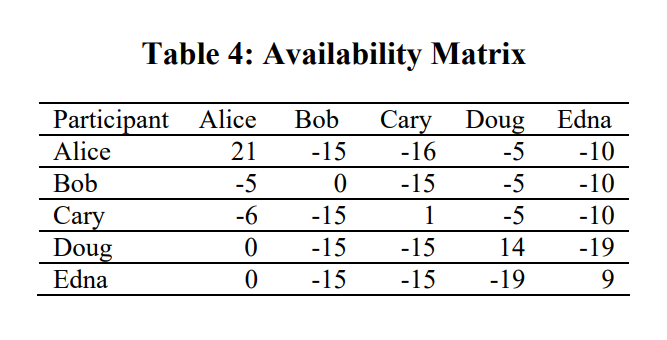
Availability Matrix (First round) 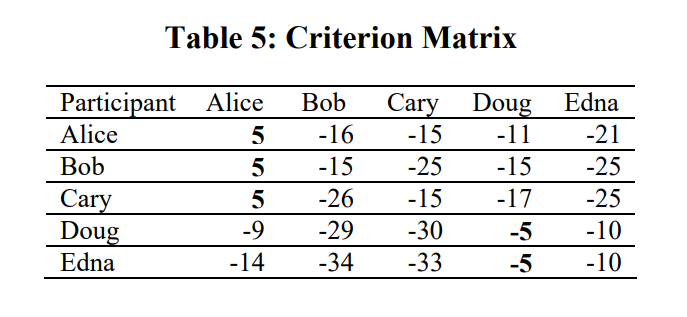
Criterion Matrix
2. Pros & Cons
Pros
- Does not need to specify the cluster number
- Allows for non-metric dissimilarities (i.e. we can have dissimilarities that don't obey the triangle inequality, or aren't symmetric)
- Providebetter stability over runs
Cons
- Similar issue as K-means: susceptible to outliers
- Affinity Propagation tends to be very slow. In practice running it on large datasets is essentially impossible without a carefully crafted and optimized implementation
3. Code Implementation
1 | from sklearn.cluster import AffinityPropagation |
Clustering: Affinity Propagation
https://criss-wang.github.io/post/blogs/unsupervised/clustering-4/